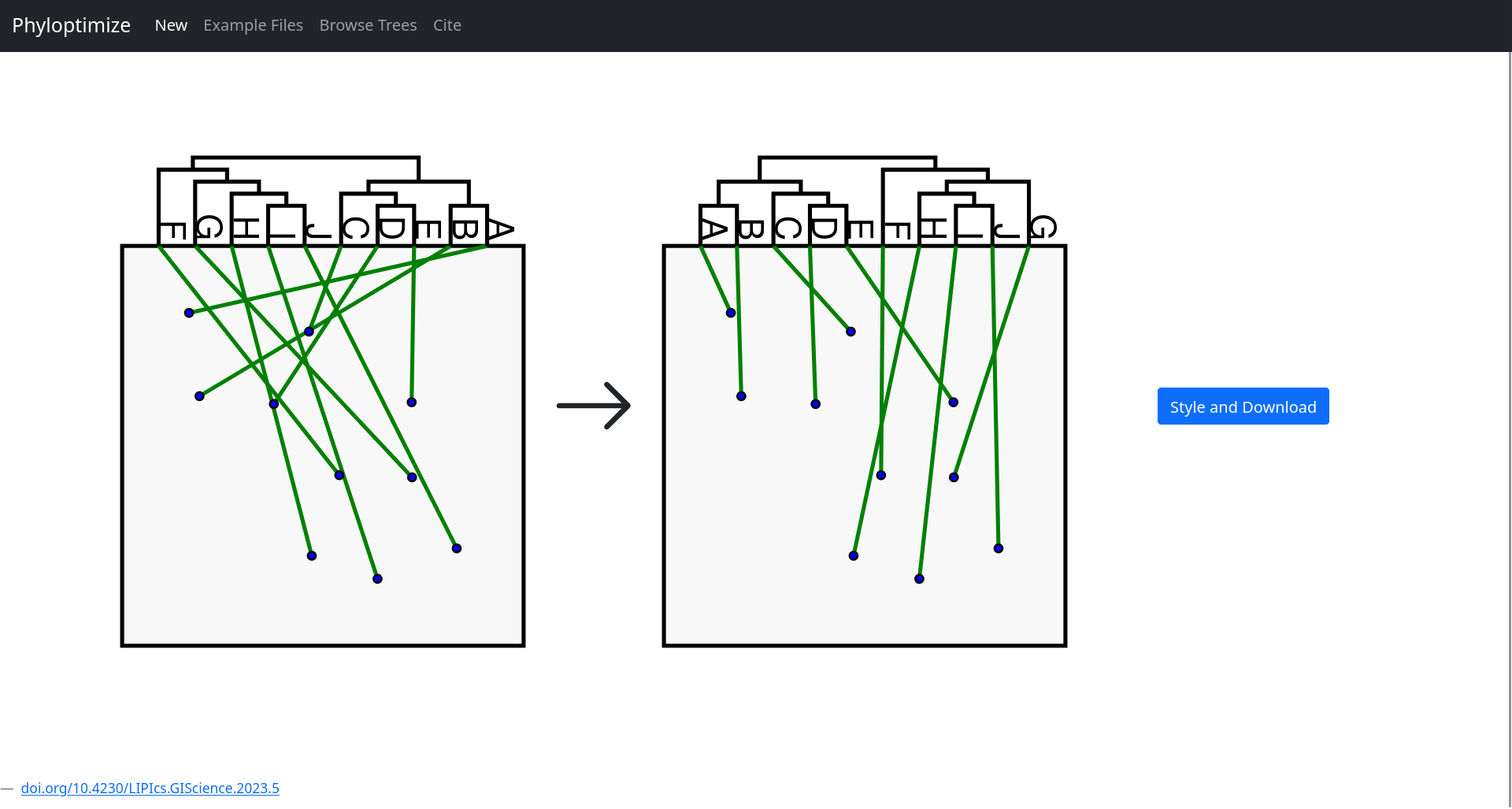
Phyloptimize was developed during my work at the Chair of Ecoinformatics at Ruhr University Bochum. It is a Python-based application that improves the readability of geophylogenetic trees by minimizing edge crossings in visualizations — a common issue when projecting such trees onto geographic maps.
The tool formulates the visualization problem as an Integer Linear Program (ILP), solved using Gurobi. To make it accessible and interactive, I designed a Flask-powered frontend where users can:
- Upload phylogenetic trees and geolocation files
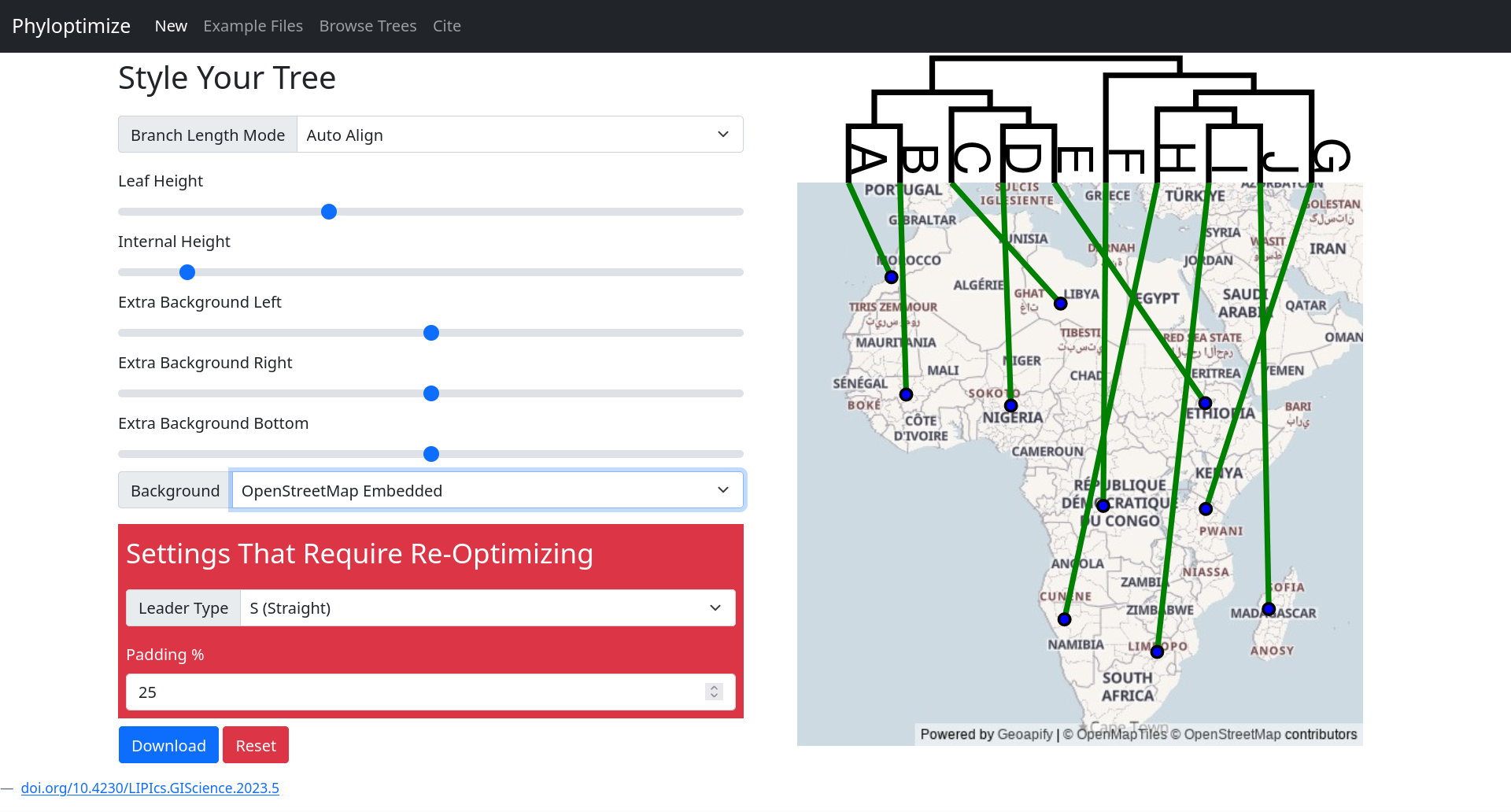
- Customize visual parameters (e.g., edge curves, leader length)
- Download optimized diagrams with static map backgrounds (via Geoapify)
Given that optimization tasks can be computationally intensive, I integrated Celery and Redis to handle these asynchronously in a separate task queue, ensuring the frontend remains responsive. The entire application is containerized using Docker, allowing scalable deployment and easier maintenance.
The underlying algorithm and results were presented in a peer-reviewed
GIScience 2023 conference publication. An open-source release is planned once development is finalized, along with public hosting for broader community use.